

The goal of this widget is to allow you to improve an alignment by forcing atoms in the embedding closer to atoms in the reference molecule. This is done by setting up an atom-atom correspondence between the two molecules (assigning atoms in the aligned structure to atoms in the reference structure) and then clicking the Refine Alignment button to run the refinement. The updated alignment will be displayed in PyMol and its important parameters (energy, shape score, and RMSD) will be displayed in the widget:
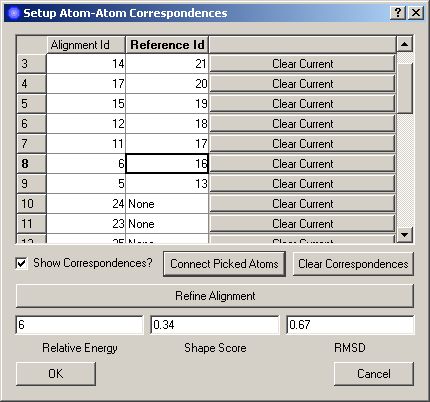
When the alignment-refinement widget first opens, a starting atom-atom correspondence will be automatically generated based on proximity of atoms in the alignment to those in the reference molecule. This correspondence is displayed in the correspondence table using atom indices and in PyMol using distance monitors to connect corresponding atoms. Clicking on a row in the correpondence table will cause the selected atom-atom correspondence to be highlighted in PyMol using atom picks (large spheres). If an atom in the alignment has no correspondence, selecting its row will highlight only that atom in PyMol.
Clicking the Clear Correspondences button will clear all current atom-atom correspondences and allow you to start afresh.
Some notes about atom-atom correspondences:
Once you are satisfied with the refined alignment, clicking the OK button will close this widget and add a new alignment to the results tree. The new alignment will be selected. Clicking the Cancel button will close the widget and discard the current refined alignment; the original alignment in the results tree will not be altered.