1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
77
78
79
80
81
82
83
84
85
86
87
88
89
90
91
92
93
94
95
96
97
98
99
100
101
102
103
104
105
106
107
108
109
110
111
112
113
114
115
116
117
118
119
120
121
122
123
124
125
126
127
128
129
130
131
132
133
134
135
136
137
138
139
140
141
142
143
144
145
146
147
148
149
150
151
152
153
154
155
156
157
158
159
160
161
162
163
164
165
166
167
168
169
170
171
172
173
174
175
176
177
178
179
180
181
182
183
184
185
186
187
188
189
190
191
192
193
194
|
[](https://maven-badges.herokuapp.com/maven-central/org.rcsb/ciftools-java)
[](https://github.com/rcsb/ciftools-java/blob/master/CHANGELOG.md)
[](https://doi.org/10.5281/zenodo.3948501)
# CIFTools
CIFTools implements reading and writing of CIF files ([specification](http://www.iucr.org/resources/cif/spec/version1.1/cifsyntax))
as well as their efficiently encoded counterpart, called BinaryCIF. The idea is to have a robust, type-safe
implementation for the handling of CIF files which does not care about the origin of the data: both conventional
text-based and binary files should be handled the same way.
## Getting Started
CIFTools is distributed by maven. To get started, append your `pom.xml` by:
```xml
<dependency>
<groupId>org.rcsb</groupId>
<artifactId>ciftools-java</artifactId>
<version>6.0.0</version>
</dependency>
```
Requires Java 11.
## File Parsing Example
```Java
class Demo {
public static void main(String[] args) {
String pdbId = "1acj";
boolean parseBinary = true;
// CIF and BinaryCIF are stored in the same data structure
// to access the data, it does not matter where and in which format the data came from
// all relevant IO operations are exposed by the CifIO class
CifFile cifFile;
if (parseBinary) {
// parse binary CIF from RCSB PDB
cifFile = CifIO.readFromURL(new URL("https://models.rcsb.org/" + pdbId + ".bcif"));
} else {
// parse CIF from RCSB PDB
cifFile = CifIO.readFromURL(new URL("https://files.rcsb.org/download/" + pdbId + ".cif"));
}
// fine-grained options are available in the CifOptions class
// access can be generic or using a specified schema - currently supports MMCIF and CIF_CORE
// you can even use a custom dictionary
MmCifFile mmCifFile = cifFile.as(StandardSchemata.MMCIF);
// get first block of CIF
MmCifBlock data = mmCifFile.getFirstBlock();
// get category with name '_atom_site' from first block - access is type-safe, all categories
// are inferred from the CIF schema
AtomSite atomSite = data.getAtomSite();
FloatColumn cartnX = atomSite.getCartnX();
// obtain entry id
String entryId = data.getEntry().getId().get(0);
System.out.println(entryId);
// calculate the average x-coordinate - #values() returns as DoubleStream as defined by the
// schema for column 'Cartn_x'
OptionalDouble averageCartnX = cartnX.values().average();
averageCartnX.ifPresent(System.out::println);
// print the last residue sequence id - this time #values() returns an IntStream
OptionalInt lastLabelSeqId = atomSite.getLabelSeqId().values().max();
lastLabelSeqId.ifPresent(System.out::println);
// print record type - or #values() may be text
Optional<String> groupPdb = data.getAtomSite().getGroupPDB().values().findFirst();
groupPdb.ifPresent(System.out::println);
}
}
```
No difference exists in the API between text-based and binary CIF files. CIF files organize data in blocks, which contain
categories (e.g. `AtomSite`), which contain columns (e.g. `CartnX`), which contain values of a particular type (e.g.
`double` values representing x-coordinates of atoms). The correct names and types for all defined categories and column
from the CIF dictionary are provided.
Just as in Mol* implementation, all parsing and decoding is done as lazily as possible. This makes it cheap to acquire
the data structure and hardly wastes any time on preparing information you will never access. In contrast to
[MMTF](https://mmtf.rcsb.org/), all data can be accessed if needed.
## Model Creation Example
```Java
class Demo {
public static void main(String[] args) {
// all builder functionality is exposed by the CifBuilder class
// again access can be generic or following a given schema
MmCifFile cifFile = CifBuilder.enterFile(StandardSchemata.MMCIF)
// create a block
.enterBlock("1EXP")
// create a category with name 'entry'
.enterEntry()
// set value of column 'id'
.enterId()
// to '1EXP'
.add("1EXP")
// leave current column
.leaveColumn()
// and category
.leaveCategory()
// create atom site category
.enterAtomSite()
// and specify some x-coordinates
.enterCartnX()
.add(1.0, -2.4, 4.5)
// values can be unknown or not specified
.markNextUnknown()
.add(-3.14, 5.0)
.leaveColumn()
// after leaving, the builder is in AtomSite again and provides column names
.enterCartnY()
.add(0.0, -1.0, 2.72)
.markNextNotPresent()
.add(42, 100)
.leaveColumn()
// leaving the builder will release the CifFile instance
.leaveCategory()
.leaveBlock()
.leaveFile();
// the created CifFile instance behaves like a parsed file and can be processed or written as needed
System.out.println(new String(CifIO.writeText(cifFile)));
System.out.println(cifFile.getFirstBlock().getEntry().getId().get(0));
cifFile.getFirstBlock()
.getAtomSite()
.getCartnX()
.values()
.forEach(System.out::println);
}
}
```
A step-wise builder is provided for the creation of `CifFile` instances. If a schema is provided, the builder is aware
of category and column names and the corresponding type described by a column (e.g. the `add` function called above is
not overloaded, but rather will only accept `String` values while in `entry.id` and only `double` values in
`atom_site.Cartn_x`.
## Read AlphaFold Model & Convert to BinaryCIF
```Java
class Demo {
public static void main(String[] args) {
String id = "AF-Q76EI6-F1-model_v4";
CifFile cifFile = CifIO.readFromURL(new URL("https://alphafold.ebi.ac.uk/files/" + id + ".cif"));
MmCifFile mmCifFile = cifFile.as(StandardSchemata.MMCIF);
// access to properties from the model-extension is provided
// print average per-residue confidence score provided by AlphaFold
System.out.println(mmCifFile.getFirstBlock()
.getMaQaMetricLocal()
.getMetricValue()
.values()
.average()
.orElseThrow());
// convert to BinaryCIF representation
byte[] output = CifIO.writeBinary(mmCifFile);
}
}
```
Computed structure models, e.g. from [AlphaFold](https://alphafold.ebi.ac.uk/), are supported. Access to categories and
columns defined by the mmCIF model extension is provided. This includes e.g. quality/confidence scores of the prediction.
Structure data can be converted to BinaryCIF files for more efficient storage & parsing of millions of files.
## Performance
The implementation can read the full PDB archive (154,015 files) in little over 2 minutes. This is achieved by lazy decoding and
parsing - all columns are decoded the first time when they are actually requested. Thus, the parsing overhead is kept
minimal. Ciftools-java combines the compression and read performance of MMTF and the convenience of the CIF format.
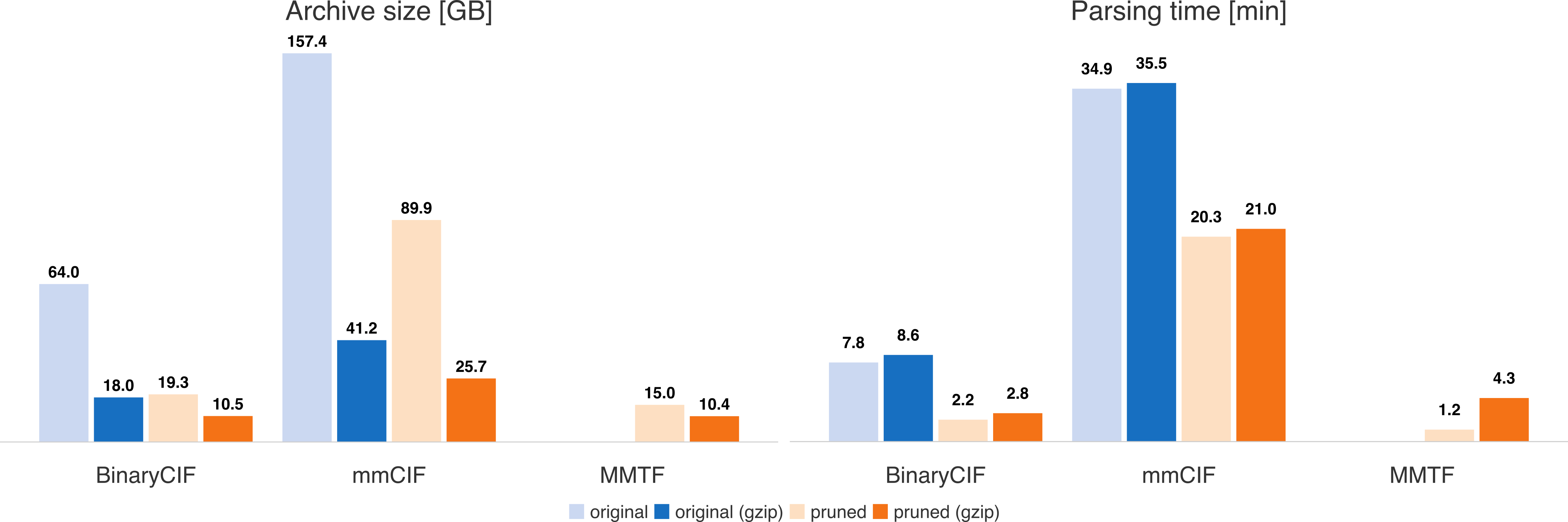
Handling gzipped files slows down parsing in most cases. The reduced files are either native MMTF files or contain a similar selection of
CIF categories (i.e. they provide primarily atomic coordinates).
## Contributions & Related Projects
- [molstar/ciftools](https://github.com/molstar/ciftools) a TypeScript/JavaScript implementation
- [molstar/BinaryCIF](https://github.com/molstar/BinaryCIF) BinaryCIF format specification
- [rcsb/py-mmcif](https://github.com/rcsb/py-mmcif) Python mmCIF Core Access Library
The implementation is based on a number of other projects, namely:
- [CIFtools.js](https://github.com/dsehnal/CIFTools.js) by David Sehnal
- [Mol*](https://molstar.github.io) by Alexander Rose and David Sehnal
- [MMTF](https://mmtf.rcsb.org/) by RCSB
## References
- Sehnal D, Bittrich S, Velankar S, Koča J, Svobodová R, Burley SK, Rose AS (2020) BinaryCIF and CIFTools—Lightweight, efficient and extensible macromolecular data management. PLoS Comput Biol 16(10): e1008247. https://doi.org/10.1371/journal.pcbi.1008247
|