1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
|
# Welcome to pycoQC v2.5.2 documentation

**PycoQC computes metrics and generates interactive QC plots for Oxford Nanopore technologies sequencing data**
PycoQC relies on the *sequencing_summary.txt* file generated by Albacore and Guppy, but if needed it can also generate a summary file from basecalled fast5 files. The package supports 1D and 1D2 runs generated with Minion, Gridion and Promethion devices, basecalled with Albacore 1.2.1+ or Guppy 2.1.3+. PycoQC is written in pure Python3. **Python 2 is not supported**. For a quick introduction see tutorial by [Tim Kahlke](https://github.com/timkahlke) available at https://timkahlke.github.io/LongRead_tutorials/QC_P.html
## Gallery

]
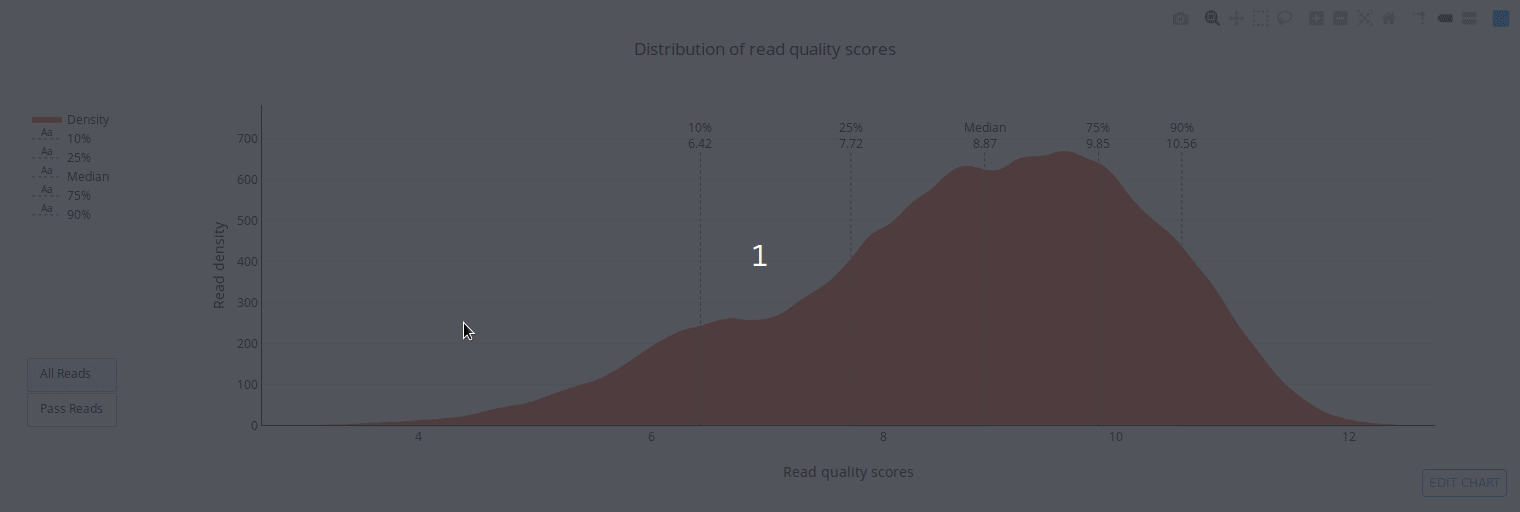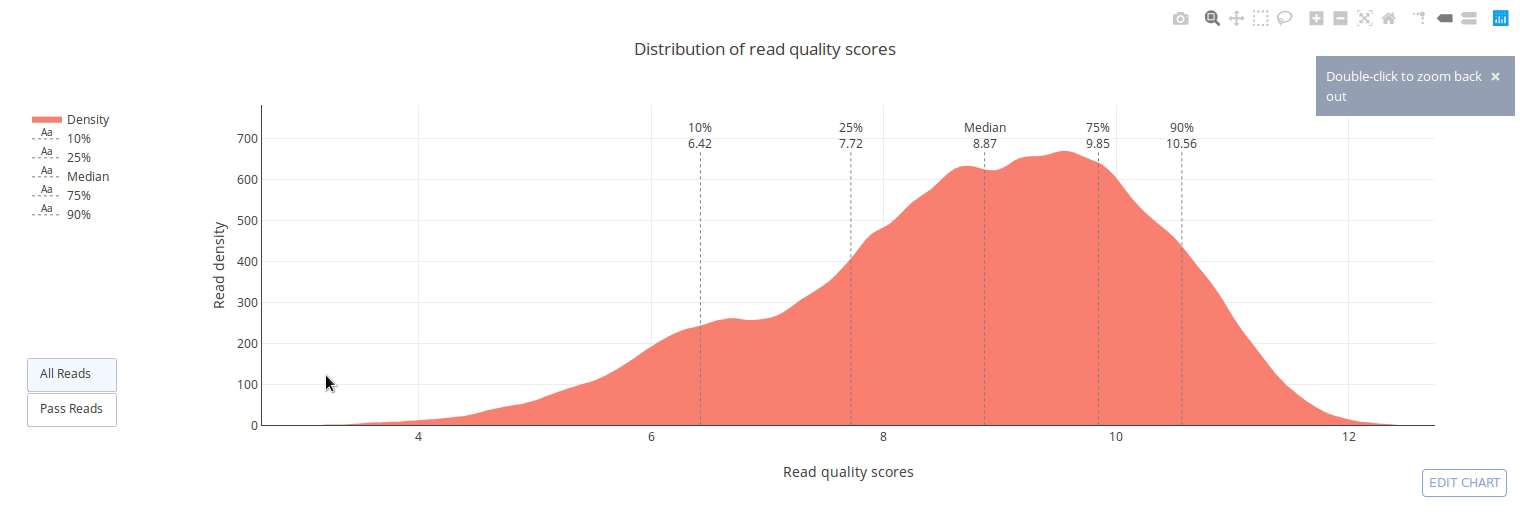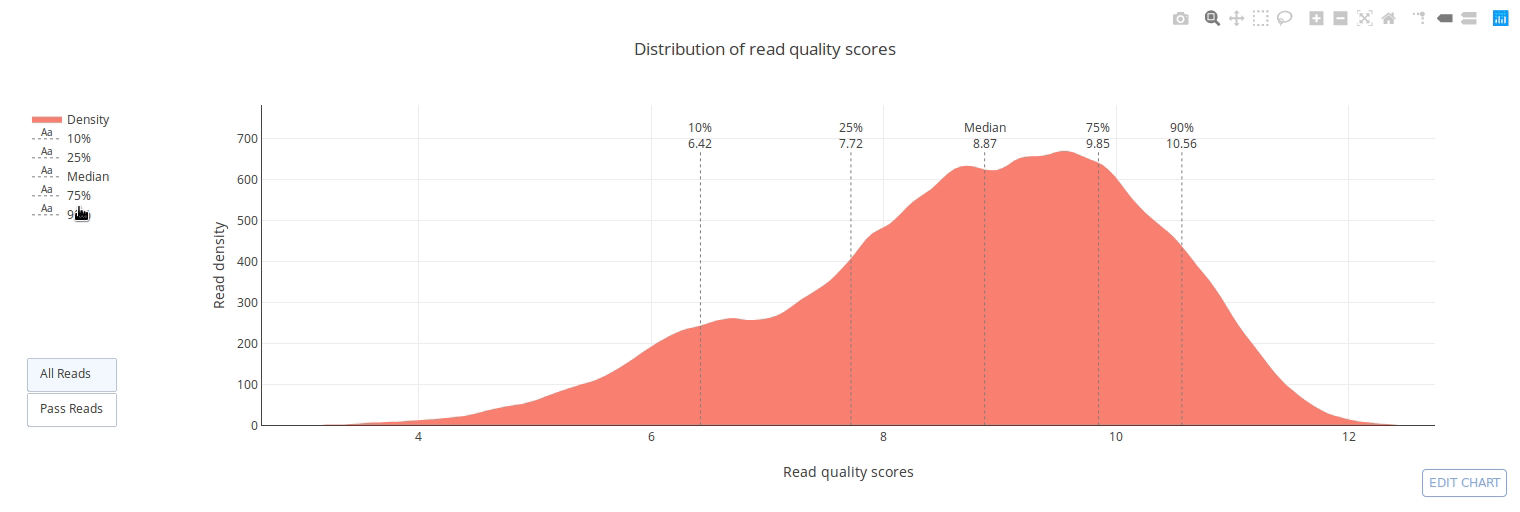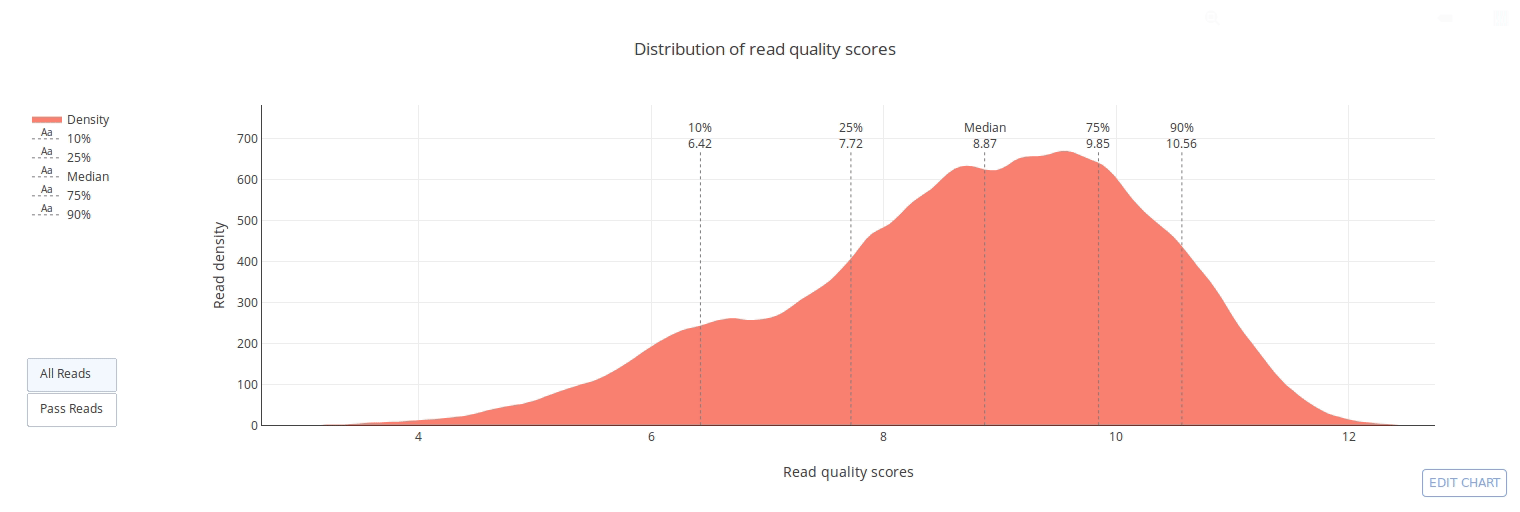]


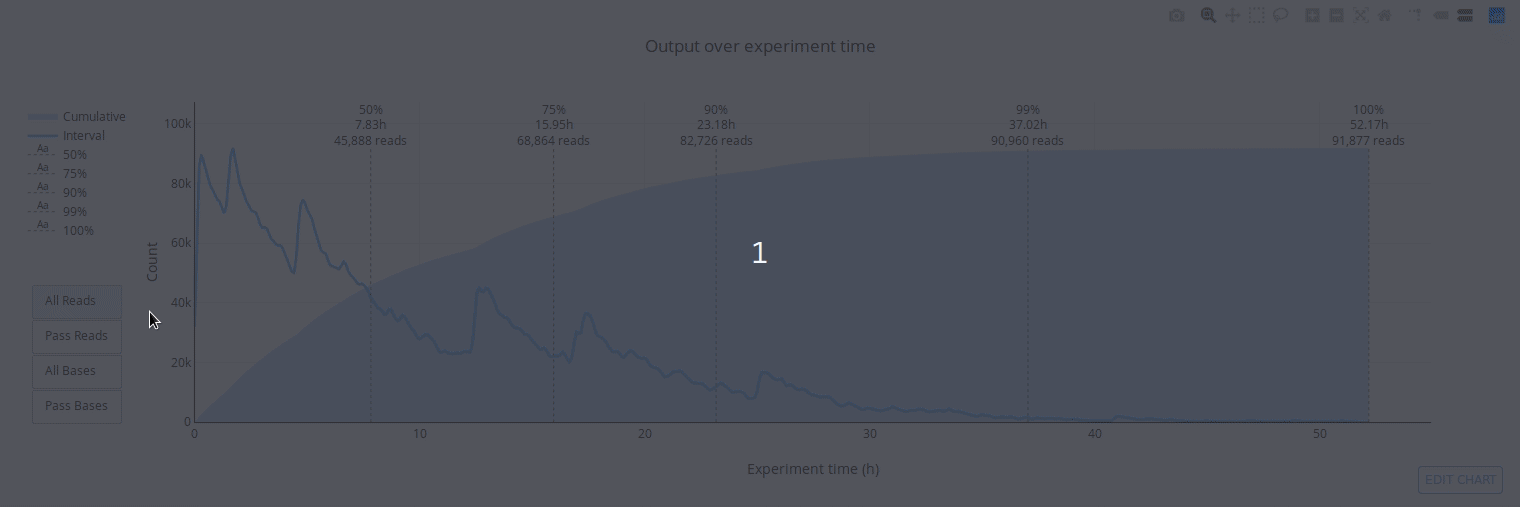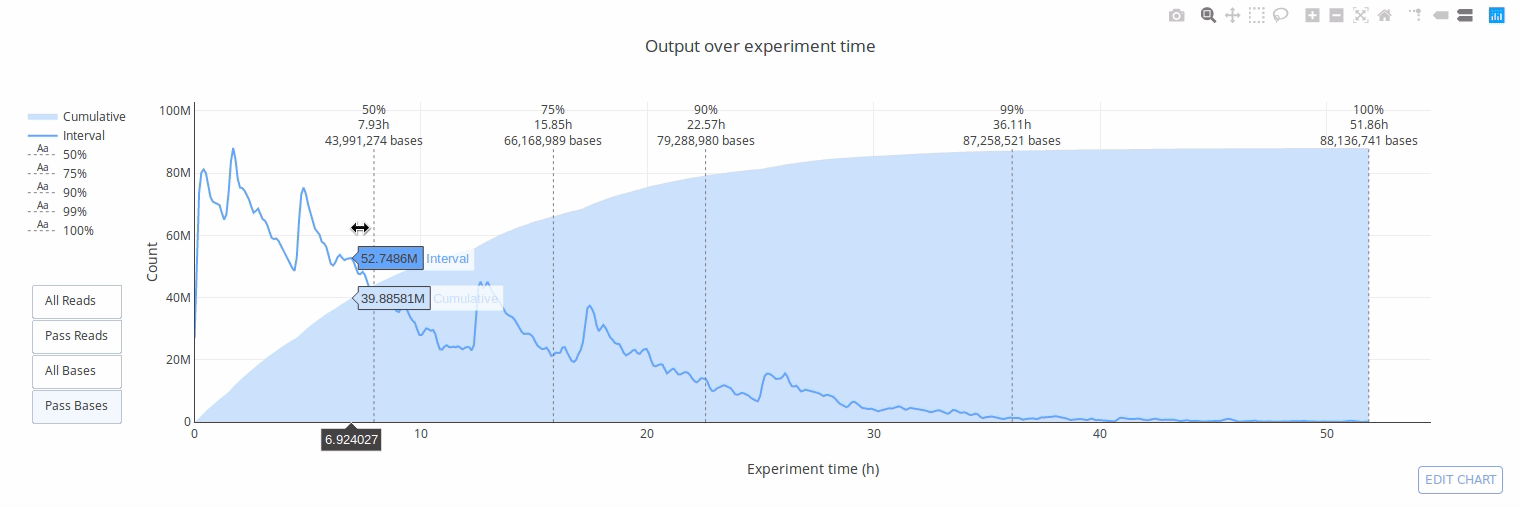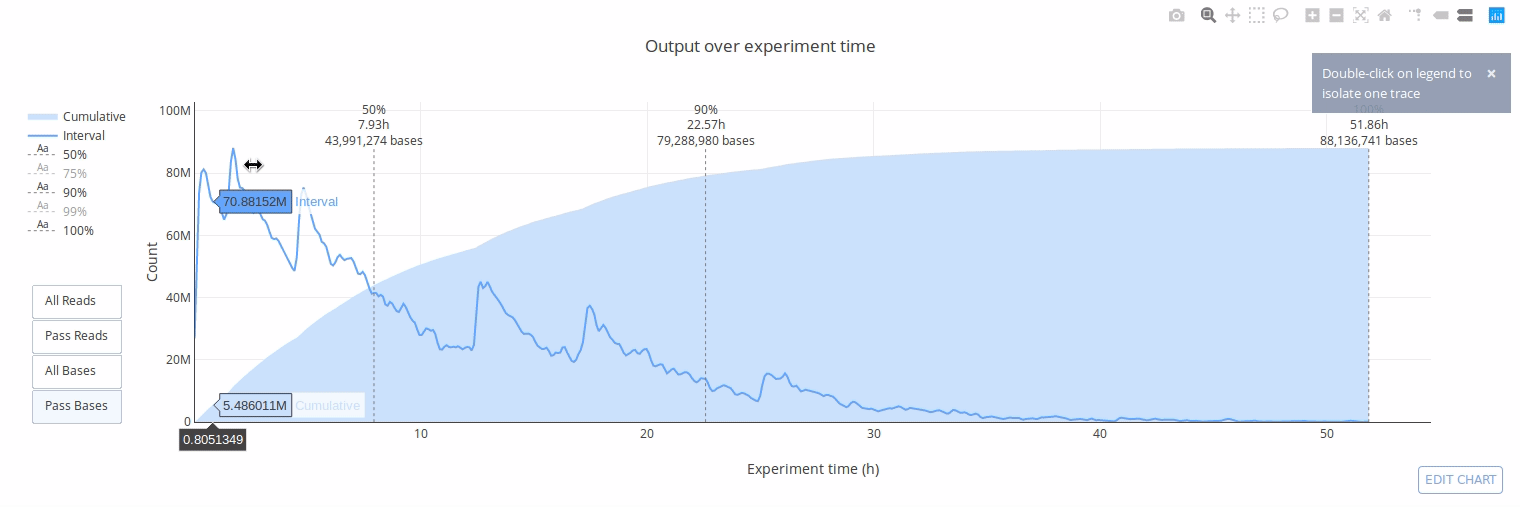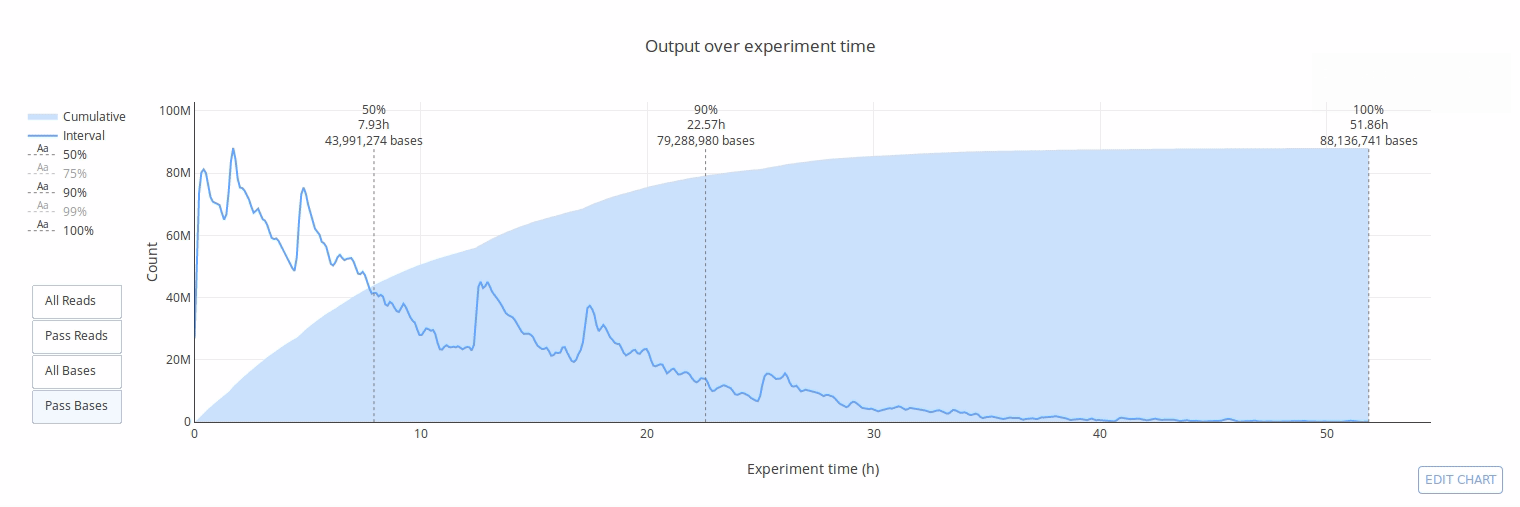








## Example HTML reports
* [Albacore_all_RNA](https://a-slide.github.io/pycoQC/pycoQC/results/Albacore_all_RNA.html)
* [Guppy-2.1.3_basecall-1D_RNA](https://a-slide.github.io/pycoQC/pycoQC/results/Guppy-2.1.3_basecall-1D_RNA.html)
* [Guppy-2.3_basecall-1D_alignment-DNA](https://a-slide.github.io/pycoQC/pycoQC/results/Guppy-2.3_basecall-1D_alignment-DNA.html)
* [Albacore-1.2.1_basecall-1D-DNA](https://a-slide.github.io/pycoQC/pycoQC/results/Albacore-1.2.1_basecall-1D-DNA.html)
* [Guppy-2.1.3_basecall-1D_DNA_barcode](https://a-slide.github.io/pycoQC/pycoQC/results/Guppy-2.1.3_basecall-1D_DNA_barcode.html)
* [Albacore-1.7.0_basecall-1D-DNA_API](https://a-slide.github.io/pycoQC/pycoQC/results/Albacore-1.7.0_basecall-1D-DNA_API.html)
* [Albacore-2.1.10_basecall-1D-DNA](https://a-slide.github.io/pycoQC/pycoQC/results/Albacore-2.1.10_basecall-1D-DNA.html)
* [Albacore-1.7.0_basecall-1D-DNA](https://a-slide.github.io/pycoQC/pycoQC/results/Albacore-1.7.0_basecall-1D-DNA.html)
## Example JSON reports
* [Guppy-2.3_basecall-1D_alignment-DNA](https://a-slide.github.io/pycoQC/pycoQC/results/Guppy-2.3_basecall-1D_alignment-DNA.json)
* [Guppy-2.1.3_basecall-1D_RNA](https://a-slide.github.io/pycoQC/pycoQC/results/Guppy-2.1.3_basecall-1D_RNA.json)
* [Albacore-1.7.0_basecall-1D-DNA_API](https://a-slide.github.io/pycoQC/pycoQC/results/Albacore-1.7.0_basecall-1D-DNA_API.json)
|