1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
77
78
79
80
81
82
83
84
85
86
87
88
89
90
91
92
93
94
95
96
97
98
99
100
101
102
103
104
105
106
107
108
109
110
111
112
|
# pycoQC v2.5.2

[](http://joss.theoj.org/papers/ea8e08dc950622bdd5d16a65649954aa)
[](https://zenodo.org/badge/latestdoi/94531811)
[](https://gitter.im/pycoQC/community?utm_source=share-link&utm_medium=link&utm_campaign=share-link)
[](https://github.com/a-slide/pycoQC/blob/master/LICENSE)
[](https://www.python.org/)
[](https://badge.fury.io/py/pycoQC)
[](https://pepy.tech/project/pycoqc)
[](https://anaconda.org/aleg/pycoqc)
[](https://anaconda.org/aleg/pycoqc)
[](http://bioconda.github.io/recipes/pycoqc/README.html)
[](https://anaconda.org/bioconda/pycoqc)
[](https://travis-ci.com/a-slide/pycoQC)
[](https://www.codacy.com/app/a-slide/pycoQC?utm_source=github.com&utm_medium=referral&utm_content=a-slide/pycoQC&utm_campaign=Badge_Grade)
---
**PycoQC computes metrics and generates interactive QC plots for Oxford Nanopore technologies sequencing data**
PycoQC relies on the *sequencing_summary.txt* file generated by Albacore and Guppy, but if needed it can also generate a summary file from basecalled fast5 files. The package supports 1D and 1D2 runs generated with Minion, Gridion and Promethion devices, basecalled with Albacore 1.2.1+ or Guppy 2.1.3+. PycoQC is written in pure Python3. **Python 2 is not supported**. For a quick introduction see tutorial by [Tim Kahlke](https://github.com/timkahlke) available at https://timkahlke.github.io/LongRead_tutorials/QC_P.html
Full documentation is available at https://adrienleger.com/pycoQC
## Gallery

]
]








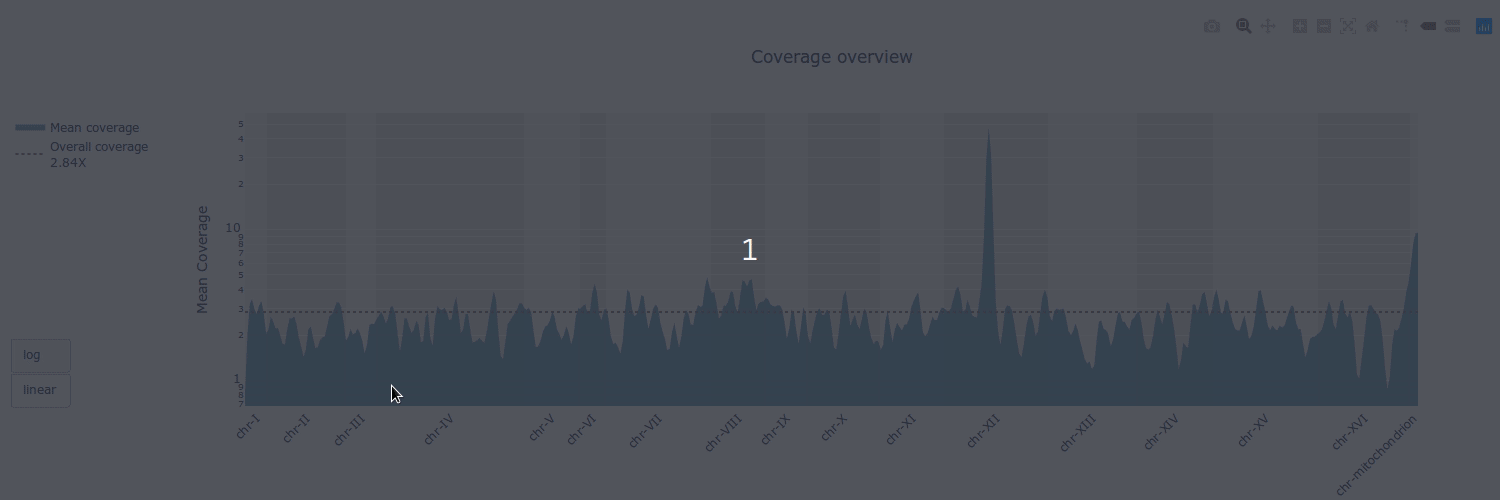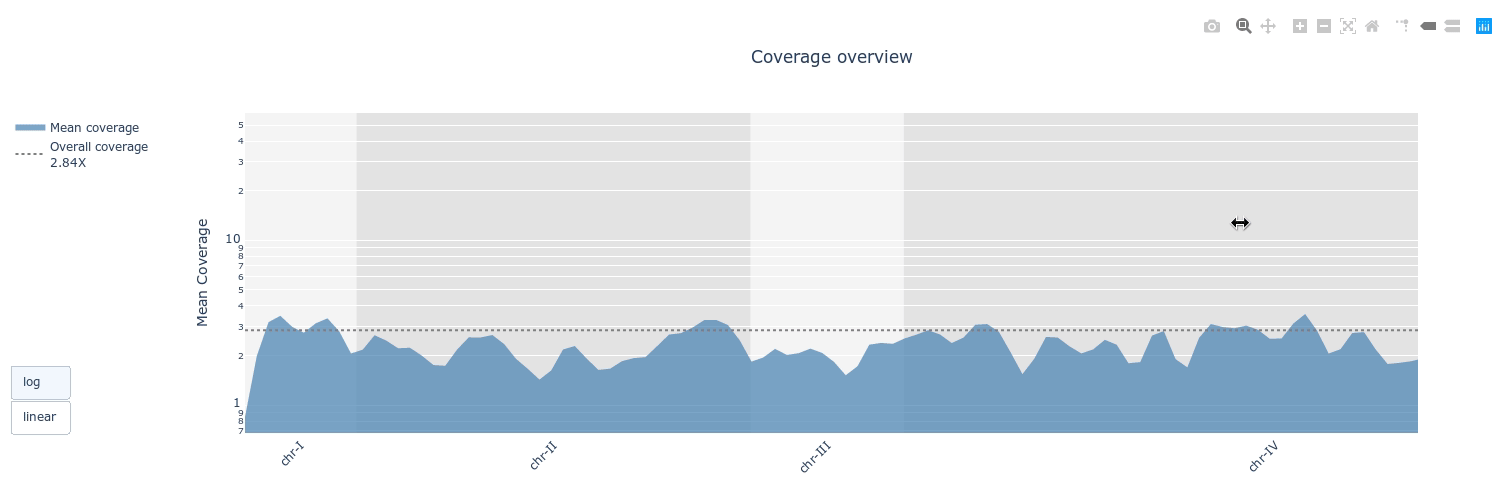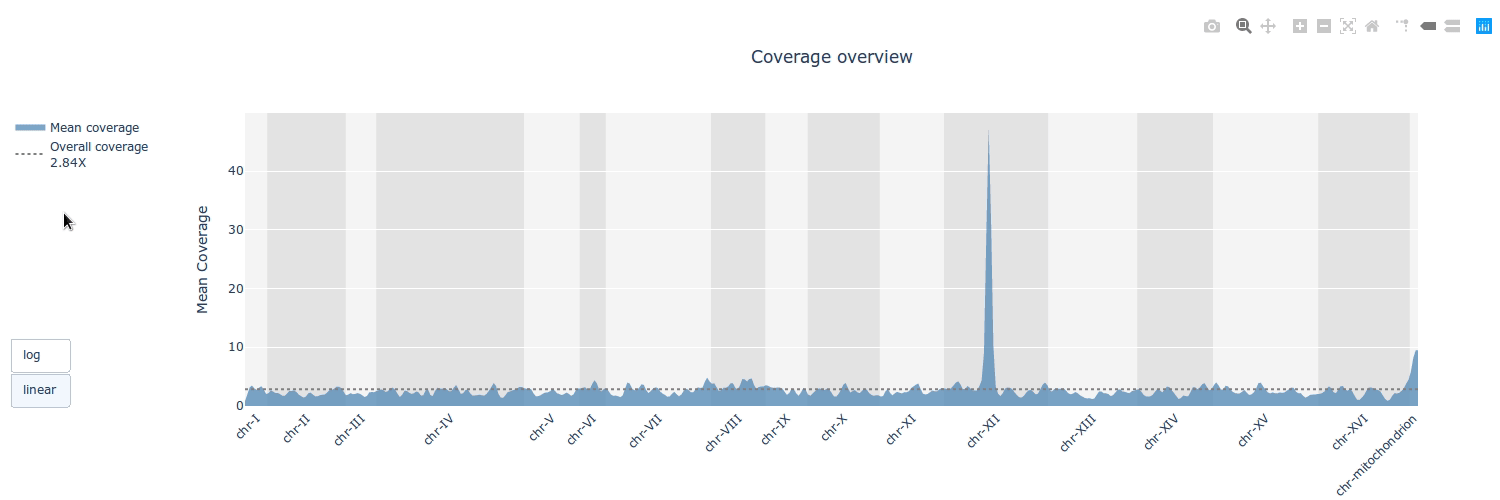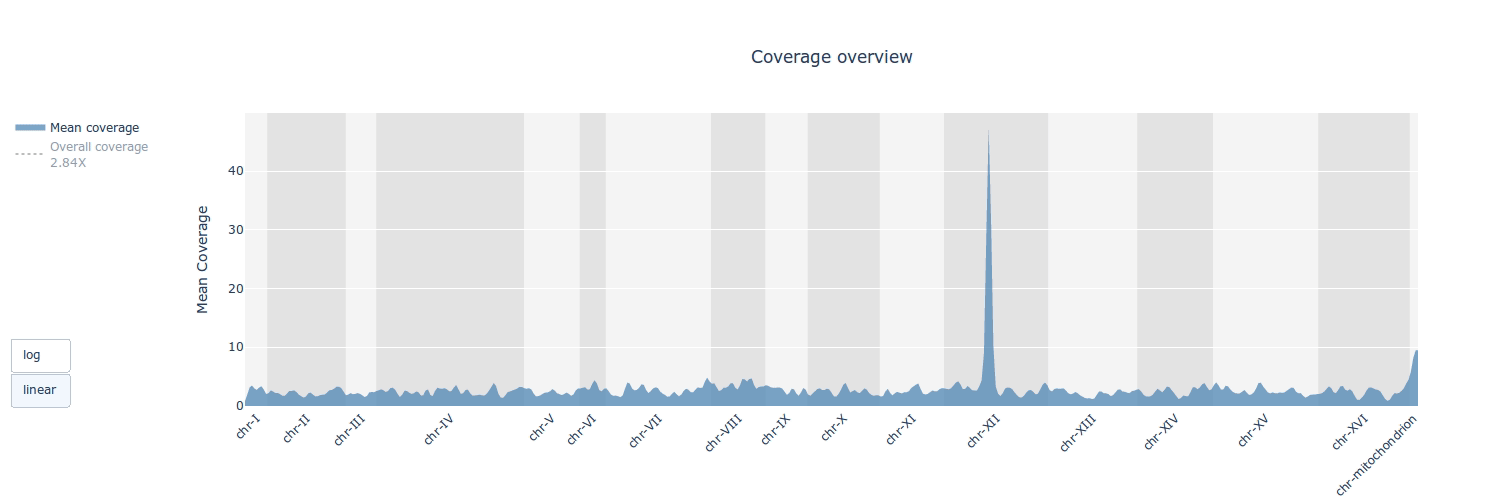


## Example HTML reports
* [Albacore_all_RNA](https://a-slide.github.io/pycoQC/pycoQC/results/Albacore_all_RNA.html)
* [Guppy-2.1.3_basecall-1D_RNA](https://a-slide.github.io/pycoQC/pycoQC/results/Guppy-2.1.3_basecall-1D_RNA.html)
* [Guppy-2.3_basecall-1D_alignment-DNA](https://a-slide.github.io/pycoQC/pycoQC/results/Guppy-2.3_basecall-1D_alignment-DNA.html)
* [Albacore-1.2.1_basecall-1D-DNA](https://a-slide.github.io/pycoQC/pycoQC/results/Albacore-1.2.1_basecall-1D-DNA.html)
* [Guppy-2.1.3_basecall-1D_DNA_barcode](https://a-slide.github.io/pycoQC/pycoQC/results/Guppy-2.1.3_basecall-1D_DNA_barcode.html)
* [Albacore-1.7.0_basecall-1D-DNA_API](https://a-slide.github.io/pycoQC/pycoQC/results/Albacore-1.7.0_basecall-1D-DNA_API.html)
* [Albacore-2.1.10_basecall-1D-DNA](https://a-slide.github.io/pycoQC/pycoQC/results/Albacore-2.1.10_basecall-1D-DNA.html)
* [Albacore-1.7.0_basecall-1D-DNA](https://a-slide.github.io/pycoQC/pycoQC/results/Albacore-1.7.0_basecall-1D-DNA.html)
## Example JSON reports
* [Guppy-2.3_basecall-1D_alignment-DNA](https://a-slide.github.io/pycoQC/pycoQC/results/Guppy-2.3_basecall-1D_alignment-DNA.json)
* [Guppy-2.1.3_basecall-1D_RNA](https://a-slide.github.io/pycoQC/pycoQC/results/Guppy-2.1.3_basecall-1D_RNA.json)
* [Albacore-1.7.0_basecall-1D-DNA_API](https://a-slide.github.io/pycoQC/pycoQC/results/Albacore-1.7.0_basecall-1D-DNA_API.json)
## Disclaimer
Please be aware that pycoQC is a research package that is still under development.
It was tested under Linux Ubuntu 16.04 and in an HPC environment running under Red Hat Enterprise 7.1.
Thank you
## Classifiers
* Development Status :: 3 - Alpha
* Intended Audience :: Science/Research
* Topic :: Scientific/Engineering :: Bio-Informatics
* License :: OSI Approved :: GNU General Public License v3 (GPLv3)
* Programming Language :: Python :: 3
## licence
GPLv3 (https://www.gnu.org/licenses/gpl-3.0.en.html)
Copyright © 2020 Adrien Leger & Tommaso Leonardi
## Authors
* Adrien Leger & Tommaso Leonardi / aleg@ebi.ac.uk / https://adrienleger.com
|