1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
77
78
79
80
81
82
83
84
85
86
87
88
89
90
91
92
93
94
95
96
97
98
99
100
101
102
103
104
105
106
107
108
109
110
111
112
113
114
115
116
117
118
119
120
121
122
123
124
125
126
127
128
129
130
131
132
133
134
135
136
137
138
139
140
141
142
143
144
145
146
147
148
149
150
151
152
153
154
155
156
157
158
159
160
161
162
163
164
165
166
167
168
169
170
171
172
173
174
175
176
177
178
179
180
181
182
183
184
185
186
187
188
189
190
191
192
193
194
195
196
197
198
199
200
201
202
203
204
205
206
207
208
209
210
211
212
213
214
215
216
217
218
219
220
221
222
223
224
225
226
227
228
229
230
231
232
233
234
235
236
237
238
239
240
241
242
243
244
245
246
247
248
249
250
251
252
253
254
255
256
257
258
259
260
261
262
263
264
265
266
267
268
269
270
271
272
273
274
275
276
277
278
279
280
281
282
283
284
285
286
287
288
289
290
291
292
293
294
295
296
297
298
299
300
301
302
303
304
305
306
307
308
309
310
311
312
313
314
315
316
317
318
319
320
321
322
323
324
325
326
327
328
329
330
331
332
333
334
335
336
337
338
339
340
341
342
343
344
345
346
347
348
349
350
351
352
353
354
355
356
357
358
359
360
361
362
363
364
365
366
367
368
369
370
371
372
373
374
375
376
377
378
379
380
381
382
383
384
385
386
387
388
389
390
391
392
393
394
395
396
397
398
399
400
401
402
403
404
405
406
407
408
409
410
411
412
413
414
415
416
417
418
419
420
421
422
423
424
425
426
427
428
429
430
431
432
433
434
435
436
437
438
439
440
441
442
443
444
445
446
447
448
449
450
451
452
453
454
455
456
457
458
459
460
461
462
463
464
465
466
467
468
469
470
471
472
473
474
475
476
|
[](https://cran.r-project.org/package=merTools)
[](https://cran.r-project.org/package=merTools)
[](https://cran.r-project.org/package=merTools)
<!-- README.md is generated from README.Rmd. Please edit that file -->
# merTools
A package for getting the most out of large multilevel models in R
by Jared E. Knowles and Carl Frederick
Working with generalized linear mixed models (GLMM) and linear mixed
models (LMM) has become increasingly easy with advances in the `lme4`
package. As we have found ourselves using these models more and more
within our work, we, the authors, have developed a set of tools for
simplifying and speeding up common tasks for interacting with `merMod`
objects from `lme4`. This package provides those tools.
## Installation
``` r
# development version
library(devtools)
install_github("jknowles/merTools")
# CRAN version
install.packages("merTools")
```
## Recent Updates
## merTools 0.6.2 (Early 2024)
- Maintenance release to fix minor issues with function documentation
- Fix \#130 by avoiding conflict with `vcov` in the `merDeriv` package
- Upgrade package test infrastructure to 3e testthat specification
## merTools 0.6.1 (Spring 2023)
- Maintenance release to keep package listed on CRAN
- Fix a small bug where parallel code path is run twice (#126)
- Update plotting functions to avoid deprecated `aes_string()` calls
(#127)
- Fix (#115) in description
- Speed up PI using @bbolker pull request (#120)
- Updated package maintainer contact information
### merTools 0.5.0
#### New Features
- `subBoot` now works with `glmerMod` objects as well
- `reMargins` a new function that allows the user to marginalize the
prediction over breaks in the distribution of random effect
distributions, see `?reMargins` and the new `reMargins` vignette
(closes \#73)
#### Bug fixes
- Fixed an issue where known convergence errors were issuing warnings
and causing the test suite to not work
- Fixed an issue where models with a random slope, no intercept, and no
fixed term were unable to be predicted (#101)
- Fixed an issue with shinyMer not working with substantive fixed
effects (#93)
### merTools 0.4.1
#### New Features
- Standard errors reported by `merModList` functions now apply the Rubin
correction for multiple imputation
#### Bug fixes
- Contribution by Alex Whitworth (@alexWhitworth) adding error checking
to plotting functions
## Shiny App and Demo
The easiest way to demo the features of this application is to use the
bundled Shiny application which launches a number of the metrics here to
aide in exploring the model. To do this:
library(merTools)
m1 <- lmer(y ~ service + lectage + studage + (1|d) + (1|s), data=InstEval)
shinyMer(m1, simData = InstEval[1:100, ]) # just try the first 100 rows of data

On the first tab, the function presents the prediction intervals for the
data selected by user which are calculated using the `predictInterval`
function within the package. This function calculates prediction
intervals quickly by sampling from the simulated distribution of the
fixed effect and random effect terms and combining these simulated
estimates to produce a distribution of predictions for each observation.
This allows prediction intervals to be generated from very large models
where the use of `bootMer` would not be feasible computationally.
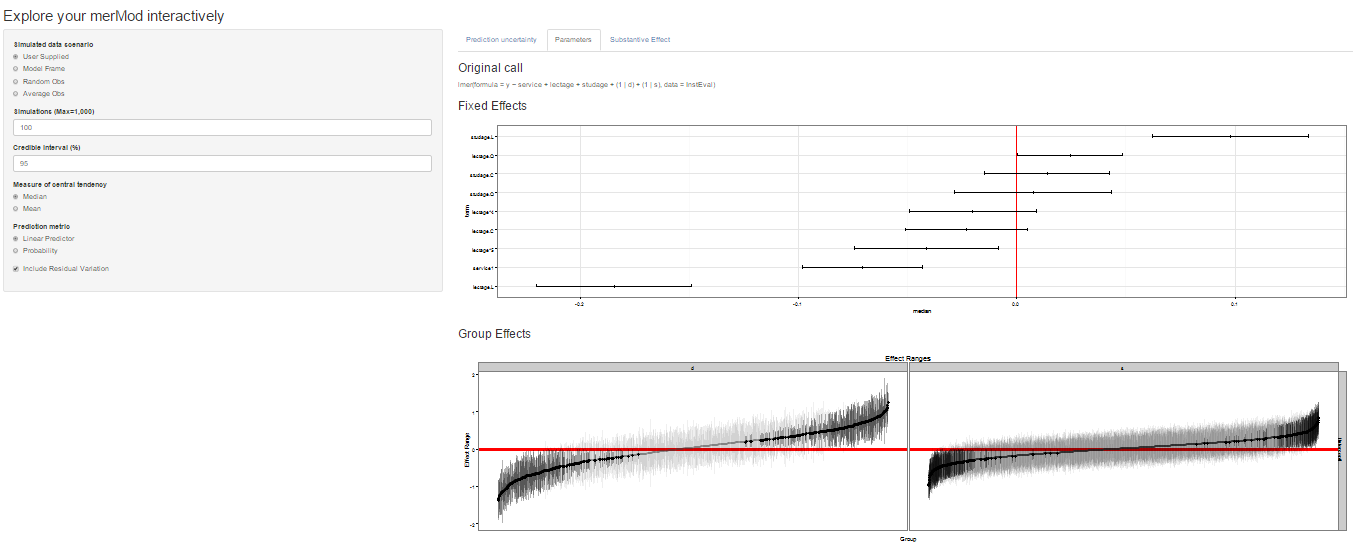
On the next tab the distribution of the fixed effect and group-level
effects is depicted on confidence interval plots. These are useful for
diagnostics and provide a way to inspect the relative magnitudes of
various parameters. This tab makes use of four related functions in
`merTools`: `FEsim`, `plotFEsim`, `REsim` and `plotREsim` which are
available to be used on their own as well.
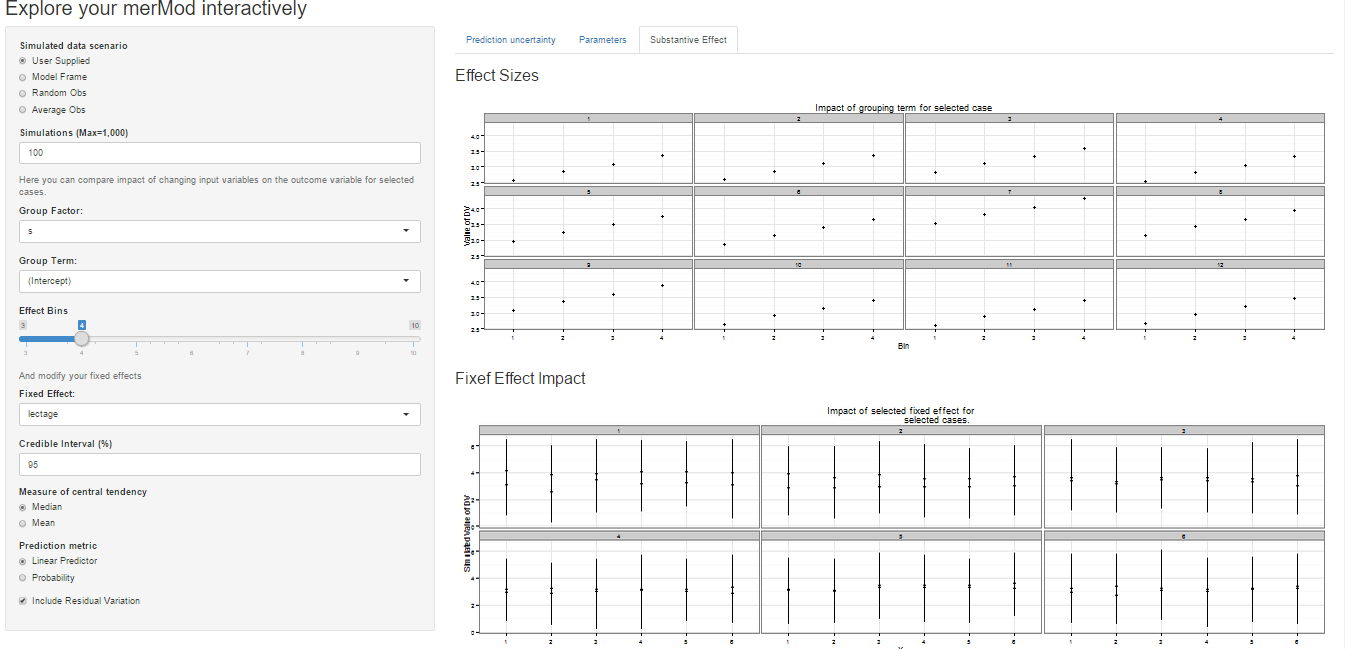
On the third tab are some convenient ways to show the influence or
magnitude of effects by leveraging the power of `predictInterval`. For
each case, up to 12, in the selected data type, the user can view the
impact of changing either one of the fixed effect or one of the grouping
level terms. Using the `REimpact` function, each case is simulated with
the model’s prediction if all else was held equal, but the observation
was moved through the distribution of the fixed effect or the random
effect term. This is plotted on the scale of the dependent variable,
which allows the user to compare the magnitude of effects across
variables, and also between models on the same data.
## Predicting
Standard prediction looks like so.
``` r
predict(m1, newdata = InstEval[1:10, ])
#> 1 2 3 4 5 6 7 8
#> 3.146337 3.165212 3.398499 3.114249 3.320686 3.252670 4.180897 3.845219
#> 9 10
#> 3.779337 3.331013
```
With `predictInterval` we obtain predictions that are more like the
standard objects produced by `lm` and `glm`:
``` r
predictInterval(m1, newdata = InstEval[1:10, ], n.sims = 500, level = 0.9,
stat = 'median')
#> fit upr lwr
#> 1 3.215698 5.302545 1.4367495
#> 2 3.155941 5.327796 1.2210140
#> 3 3.374129 5.287901 1.4875231
#> 4 3.101672 5.183841 0.9248584
#> 5 3.299367 5.298370 1.3287058
#> 6 3.147238 5.368311 1.1132248
#> 7 4.155194 6.273147 2.2167207
#> 8 3.873493 5.705669 1.9152401
#> 9 3.740978 5.737517 2.0454222
#> 10 3.291242 5.297614 1.2375007
```
Note that `predictInterval` is slower because it is computing
simulations. It can also return all of the simulated `yhat` values as an
attribute to the predict object itself.
`predictInterval` uses the `sim` function from the `arm` package heavily
to draw the distributions of the parameters of the model. It then
combines these simulated values to create a distribution of the `yhat`
for each observation.
### Inspecting the Prediction Components
We can also explore the components of the prediction interval by asking
`predictInterval` to return specific components of the prediction
interval.
``` r
predictInterval(m1, newdata = InstEval[1:10, ], n.sims = 200, level = 0.9,
stat = 'median', which = "all")
#> effect fit upr lwr obs
#> 1 combined 3.35554348 5.217964 1.615782 1
#> 2 combined 3.21487934 5.327824 1.114338 2
#> 3 combined 3.44493242 5.474256 1.809136 3
#> 4 combined 3.24123655 4.838427 1.272174 4
#> 5 combined 3.20539661 5.367651 1.068128 5
#> 6 combined 3.54335144 5.481756 1.585809 6
#> 7 combined 4.23212790 6.267669 2.284923 7
#> 8 combined 4.05055116 5.684968 1.931558 8
#> 9 combined 3.84266853 5.492163 2.091312 9
#> 10 combined 3.24121727 5.183680 1.196101 10
#> 11 s -0.02342248 1.948494 -1.691035 1
#> 12 s 0.04148408 2.091467 -1.782386 2
#> 13 s 0.04477028 2.087629 -2.144621 3
#> 14 s 0.26160482 2.114509 -1.733429 4
#> 15 s -0.10803386 1.714535 -1.982283 5
#> 16 s -0.04962613 1.916212 -1.909187 6
#> 17 s 0.24916111 2.001528 -1.628554 7
#> 18 s 0.19640074 2.070513 -1.473660 8
#> 19 s 0.27031215 2.119763 -1.643120 9
#> 20 s 0.13772544 2.313012 -1.855489 10
#> 21 d -0.32196201 1.357316 -2.397083 1
#> 22 d -0.29691477 1.422141 -2.662141 2
#> 23 d 0.24828667 1.782181 -1.987563 3
#> 24 d -0.37893052 1.471225 -2.350781 4
#> 25 d 0.02142086 2.172075 -2.148417 5
#> 26 d 0.07926221 2.003462 -1.677765 6
#> 27 d 0.76480967 2.767889 -1.274501 7
#> 28 d 0.08757337 2.374201 -1.958689 8
#> 29 d 0.25289032 2.083732 -1.376630 9
#> 30 d -0.17775160 1.601744 -2.115104 10
#> 31 fixed 3.16750528 5.010517 1.371678 1
#> 32 fixed 3.21493166 5.246672 1.074857 2
#> 33 fixed 3.36233628 5.581696 1.474776 3
#> 34 fixed 3.17926915 5.107315 1.621278 4
#> 35 fixed 3.16562882 5.136197 1.156010 5
#> 36 fixed 3.15944014 5.114967 1.506315 6
#> 37 fixed 3.32101367 5.149819 1.407884 7
#> 38 fixed 3.34020282 5.189215 1.651446 8
#> 39 fixed 3.17901802 5.000429 1.132874 9
#> 40 fixed 3.41100236 5.207451 1.555844 10
```
This can lead to some useful plotting:
``` r
library(ggplot2)
#> Warning: package 'ggplot2' was built under R version 4.3.2
plotdf <- predictInterval(m1, newdata = InstEval[1:10, ], n.sims = 2000,
level = 0.9, stat = 'median', which = "all",
include.resid.var = FALSE)
plotdfb <- predictInterval(m1, newdata = InstEval[1:10, ], n.sims = 2000,
level = 0.9, stat = 'median', which = "all",
include.resid.var = TRUE)
plotdf <- dplyr::bind_rows(plotdf, plotdfb, .id = "residVar")
plotdf$residVar <- ifelse(plotdf$residVar == 1, "No Model Variance",
"Model Variance")
ggplot(plotdf, aes(x = obs, y = fit, ymin = lwr, ymax = upr)) +
geom_pointrange() +
geom_hline(yintercept = 0, color = I("red"), size = 1.1) +
scale_x_continuous(breaks = c(1, 10)) +
facet_grid(residVar~effect) + theme_bw()
#> Warning: Using `size` aesthetic for lines was deprecated in ggplot2 3.4.0.
#> ℹ Please use `linewidth` instead.
#> This warning is displayed once every 8 hours.
#> Call `lifecycle::last_lifecycle_warnings()` to see where this warning was
#> generated.
```
<!-- -->
We can also investigate the makeup of the prediction for each
observation.
``` r
ggplot(plotdf[plotdf$obs < 6,],
aes(x = effect, y = fit, ymin = lwr, ymax = upr)) +
geom_pointrange() +
geom_hline(yintercept = 0, color = I("red"), size = 1.1) +
facet_grid(residVar~obs) + theme_bw()
```
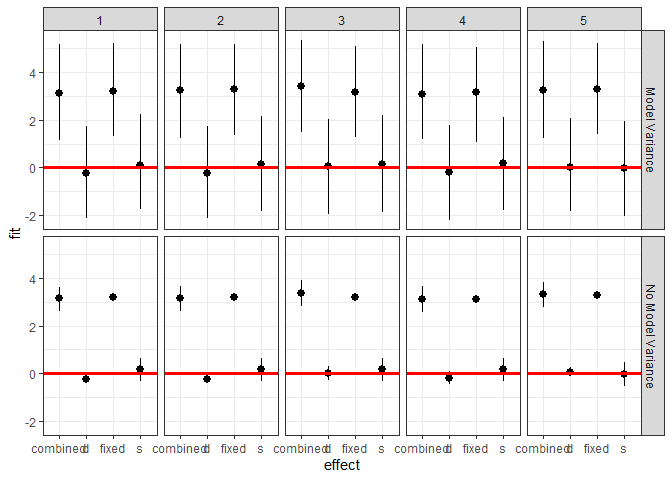<!-- -->
## Plotting
`merTools` also provides functionality for inspecting `merMod` objects
visually. The easiest are getting the posterior distributions of both
fixed and random effect parameters.
``` r
feSims <- FEsim(m1, n.sims = 100)
head(feSims)
#> term mean median sd
#> 1 (Intercept) 3.22469489 3.22427807 0.01767782
#> 2 service1 -0.07112136 -0.07190156 0.01382577
#> 3 lectage.L -0.18681442 -0.18833173 0.01617233
#> 4 lectage.Q 0.02208290 0.02252175 0.01259911
#> 5 lectage.C -0.02656846 -0.02507924 0.01099316
#> 6 lectage^4 -0.02165334 -0.02073531 0.01355675
```
And we can also plot this:
``` r
plotFEsim(FEsim(m1, n.sims = 100), level = 0.9, stat = 'median', intercept = FALSE)
```
<!-- -->
We can also quickly make caterpillar plots for the random-effect terms:
``` r
reSims <- REsim(m1, n.sims = 100)
head(reSims)
#> groupFctr groupID term mean median sd
#> 1 s 1 (Intercept) 0.21300834 0.22465501 0.3266393
#> 2 s 2 (Intercept) -0.04278168 -0.08474934 0.3164631
#> 3 s 3 (Intercept) 0.41998629 0.42229608 0.2865010
#> 4 s 4 (Intercept) 0.28818926 0.28746523 0.3019460
#> 5 s 5 (Intercept) 0.05833502 0.03342223 0.3539464
#> 6 s 6 (Intercept) 0.08108947 0.08478943 0.2049882
```
``` r
plotREsim(REsim(m1, n.sims = 100), stat = 'median', sd = TRUE)
```
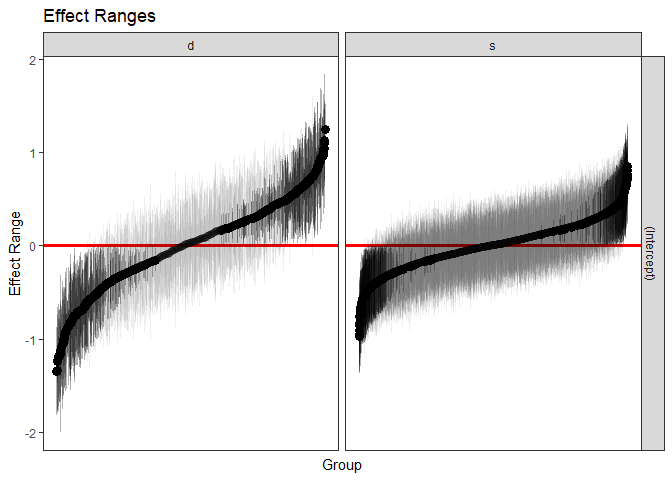<!-- -->
Note that `plotREsim` highlights group levels that have a simulated
distribution that does not overlap 0 – these appear darker. The lighter
bars represent grouping levels that are not distinguishable from 0 in
the data.
Sometimes the random effects can be hard to interpret and not all of
them are meaningfully different from zero. To help with this `merTools`
provides the `expectedRank` function, which provides the percentile
ranks for the observed groups in the random effect distribution taking
into account both the magnitude and uncertainty of the estimated effect
for each group.
``` r
ranks <- expectedRank(m1, groupFctr = "d")
head(ranks)
#> groupFctr groupLevel term estimate std.error ER pctER
#> 2 d 1 Intercept 0.3944919 0.08665152 835.3005 74
#> 3 d 6 Intercept -0.4428949 0.03901988 239.5363 21
#> 4 d 7 Intercept 0.6562681 0.03717200 997.3569 88
#> 5 d 8 Intercept -0.6430680 0.02210017 138.3445 12
#> 6 d 12 Intercept 0.1902940 0.04024063 702.3410 62
#> 7 d 13 Intercept 0.2497464 0.03216255 750.0174 66
```
A nice features `expectedRank` is that you can return the expected rank
for all factors simultaneously and use them:
``` r
ranks <- expectedRank(m1)
head(ranks)
#> groupFctr groupLevel term estimate std.error ER pctER
#> 2 s 1 Intercept 0.16732800 0.08165665 1931.570 65
#> 3 s 2 Intercept -0.04409538 0.09234250 1368.160 46
#> 4 s 3 Intercept 0.30382219 0.05204082 2309.941 78
#> 5 s 4 Intercept 0.24756175 0.06641699 2151.828 72
#> 6 s 5 Intercept 0.05232329 0.08174130 1627.693 55
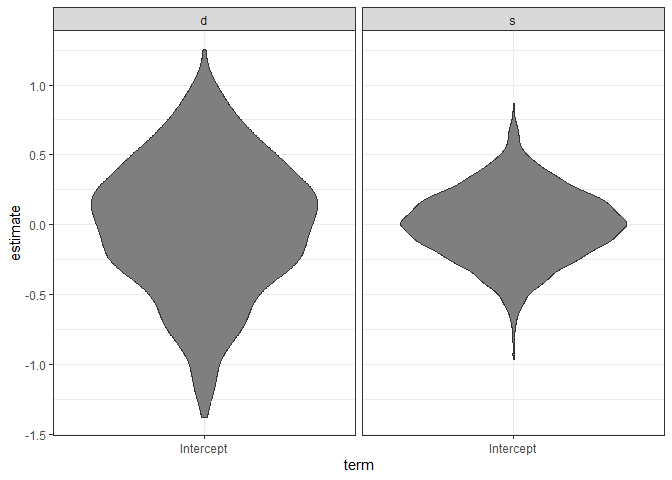
#> 7 s 6 Intercept 0.10191653 0.06648394 1772.548 60
ggplot(ranks, aes(x = term, y = estimate)) +
geom_violin(fill = "gray50") + facet_wrap(~groupFctr) +
theme_bw()
```
<!-- -->
## Effect Simulation
It can still be difficult to interpret the results of LMM and GLMM
models, especially the relative influence of varying parameters on the
predicted outcome. This is where the `REimpact` and the `wiggle`
functions in `merTools` can be handy.
``` r
impSim <- REimpact(m1, InstEval[7, ], groupFctr = "d", breaks = 5,
n.sims = 300, level = 0.9)
#> Warning: executing %dopar% sequentially: no parallel backend registered
impSim
#> case bin AvgFit AvgFitSE nobs
#> 1 1 1 2.775244 2.889334e-04 193
#> 2 1 2 3.251612 5.378474e-05 240
#> 3 1 3 3.535261 5.204943e-05 254
#> 4 1 4 3.834013 6.310989e-05 265
#> 5 1 5 4.220246 1.859937e-04 176
```
The result of `REimpact` shows the change in the `yhat` as the case we
supplied to `newdata` is moved from the first to the fifth quintile in
terms of the magnitude of the group factor coefficient. We can see here
that the individual professor effect has a strong impact on the outcome
variable. This can be shown graphically as well:
``` r
ggplot(impSim, aes(x = factor(bin), y = AvgFit, ymin = AvgFit - 1.96*AvgFitSE,
ymax = AvgFit + 1.96*AvgFitSE)) +
geom_pointrange() + theme_bw() + labs(x = "Bin of `d` term", y = "Predicted Fit")
```
<!-- -->
Here the standard error is a bit different – it is the weighted standard
error of the mean effect within the bin. It does not take into account
the variability within the effects of each observation in the bin –
accounting for this variation will be a future addition to `merTools`.
## Explore Substantive Impacts
Another feature of `merTools` is the ability to easily generate
hypothetical scenarios to explore the predicted outcomes of a `merMod`
object and understand what the model is saying in terms of the outcome
variable.
Let’s take the case where we want to explore the impact of a model with
an interaction term between a category and a continuous predictor.
First, we fit a model with interactions:
``` r
data(VerbAgg)
fmVA <- glmer(r2 ~ (Anger + Gender + btype + situ)^2 +
(1|id) + (1|item), family = binomial,
data = VerbAgg)
#> Warning in checkConv(attr(opt, "derivs"), opt$par, ctrl = control$checkConv, :
#> Model failed to converge with max|grad| = 0.0543724 (tol = 0.002, component 1)
```
Now we prep the data using the `draw` function in `merTools`. Here we
draw the average observation from the model frame. We then `wiggle` the
data by expanding the dataframe to include the same observation repeated
but with different values of the variable specified by the `var`
parameter. Here, we expand the dataset to all values of `btype`, `situ`,
and `Anger` subsequently.
``` r
# Select the average case
newData <- draw(fmVA, type = "average")
newData <- wiggle(newData, varlist = "btype",
valueslist = list(unique(VerbAgg$btype)))
newData <- wiggle(newData, var = "situ",
valueslist = list(unique(VerbAgg$situ)))
newData <- wiggle(newData, var = "Anger",
valueslist = list(unique(VerbAgg$Anger)))
head(newData, 10)
#> r2 Anger Gender btype situ id item
#> 1 N 20 F curse other 5 S3WantCurse
#> 2 N 20 F scold other 5 S3WantCurse
#> 3 N 20 F shout other 5 S3WantCurse
#> 4 N 20 F curse self 5 S3WantCurse
#> 5 N 20 F scold self 5 S3WantCurse
#> 6 N 20 F shout self 5 S3WantCurse
#> 7 N 11 F curse other 5 S3WantCurse
#> 8 N 11 F scold other 5 S3WantCurse
#> 9 N 11 F shout other 5 S3WantCurse
#> 10 N 11 F curse self 5 S3WantCurse
```
The next step is familiar – we simply pass this new dataset to
`predictInterval` in order to generate predictions for these
counterfactuals. Then we plot the predicted values against the
continuous variable, `Anger`, and facet and group on the two categorical
variables `situ` and `btype` respectively.
``` r
plotdf <- predictInterval(fmVA, newdata = newData, type = "probability",
stat = "median", n.sims = 1000)
plotdf <- cbind(plotdf, newData)
ggplot(plotdf, aes(y = fit, x = Anger, color = btype, group = btype)) +
geom_point() + geom_smooth(aes(color = btype), method = "lm") +
facet_wrap(~situ) + theme_bw() +
labs(y = "Predicted Probability")
#> `geom_smooth()` using formula = 'y ~ x'
```
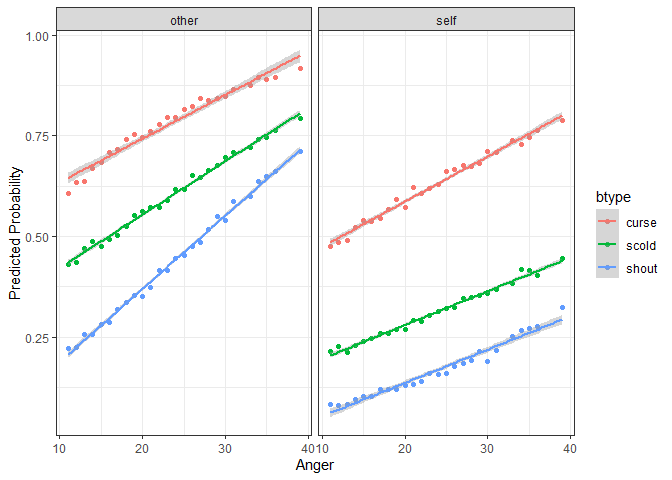<!-- -->
## Marginalizing Random Effects
``` r
# get cases
case_idx <- sample(1:nrow(VerbAgg), 10)
mfx <- REmargins(fmVA, newdata = VerbAgg[case_idx,], breaks = 4, groupFctr = "item",
type = "probability")
ggplot(mfx, aes(y = fit_combined, x = breaks, group = case)) +
geom_point() + geom_line() +
theme_bw() +
scale_y_continuous(breaks = 1:10/10, limits = c(0, 1)) +
coord_cartesian(expand = FALSE) +
labs(x = "Quartile of item random effect Intercept for term 'item'",
y = "Predicted Probability",
title = "Simulated Effect of Item Intercept on Predicted Probability for 10 Random Cases")
```
<!-- -->
|