1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
77
78
79
80
81
82
83
84
85
86
87
88
89
90
91
92
93
94
95
96
97
98
99
100
101
102
103
104
105
106
107
108
109
110
111
112
113
114
115
116
117
118
119
120
121
122
123
124
125
126
127
128
129
130
131
132
133
134
135
136
137
138
139
140
141
142
143
144
145
146
147
148
149
150
151
152
153
|
# seriation
This software is an efficient implementation of the [Seriation](http://www.jstatsoft.org/v25/i03) problem which 'finds a suitable linear order for a set of objects'. It has been used to order a network of proteins such that 'related' nodes are closer in the other.
## Authors
The Seriation Package was developed by [Felipe Kuentzer](http://lattes.cnpq.br/1979213773480902), in collaboration with
Douglas G. Ávila, Alexandre Pereira, Gabriel Perrone, Samoel da Silva, [Alexandre Amory](http://lattes.cnpq.br/2609000874577720), and [Rita de Almeida](http://lattes.cnpq.br/4672766298301524).
**Contact information**: Alexandre Amory (*alexandre.amory at pucrs.br*)
## Inputs
The input file is a textual file describing an undirected network of nodes (in our examples the nodes are protein names). Example:
<pre>
L7007 L7008
L7008 L7007
L7010 L7011
L7011 L7010
L7014 L7015
L7015 L7014
L7017 Z1275
</pre>
In the tab Files you can find networks for different species such as [Escherichia coli](data/Escherichia_coli.dat), [Mus musculus](data/Mus_musculus.dat), [Saccharomyces cerevisiae](data/Saccharomyces_cerevisiae.dat), [Homo sapiens](data/Homo_sapiens.dat), among others.
## Outputs
The output is a text file with the order of the network nodes. Example:
<pre>
Protein dim1
Z5822 0
Z5823 1
Z2911 2
Z2910 3
Z2909 4
Z4123 5
Z4124 6
Z3105 7
Z3106 8
...
</pre>
The following image represents the Homo sapiens network with a *random ordering*.
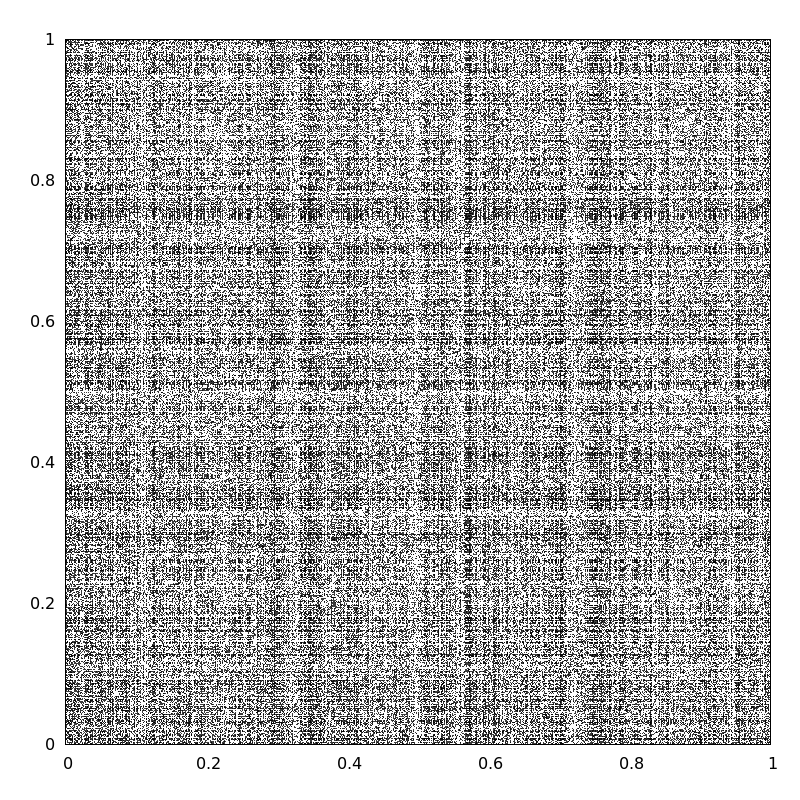
The next image represents the Homo sapiens network **'seriated'**.

## Download and Compilation
The Seriation Package is developed in C and tested on Ubuntu 14.04.
* Download the package ("amd64":/redmine/attachments/download/360/cfm-seriation_1.0-1_amd64.deb) or ("i386":/redmine/attachments/download/361/cfm-seriation_1.0-1_i386.deb).
* Is recommended to update your packages before the instalation:
> * sudo apt-get install update
* To install, you can double-click it or execute:
> * sudo dpkg -i cfm-seriation_1.0-1_amd64.deb
> * In case of missing dependencies, try: sudo apt-get install -f
* To unistall:
> * sudo dpkg -r cfm-seriation
* this distribution has the following files
<pre>
/usr/share/cfm-seriation/bin/ Executable file
/usr/share/cfm-seriation/etc/ Auxiliar used to plot charts with GNUPLOT
/usr/share/cfm-seriation/data/ Biological input networks
/usr/share/cfm-seriation/src/ Source code in C
</pre>
## How to Use
type 'cfm-seriation' to show the options:
<pre>
cfm-seriation
Usage: cfm-seriation [OPTION...]
Seriation Parameters:
f=[NETWORK FILE].dat Network file path name
o=[ORDER FILE].dat Apply initial order
i=[INTERVAL] Number of isothermal steps
m=[STEPS] Number of steps
c=[FACTOR] Cooling factor
a=[ALPHA] Alpha value
p=[PERCENTUAL] Percentual energy for initial temperature
s=[SEDD] Random seed
P Plot graphs
v Generate video
</pre>
type 'cfm-seriation f=/usr/share/cfm-seriation/data/Homo_sapiens.dat m=3000 P' to execute the seriation. This process can take about 12 minutes, depending on the CPU.
In case you want a video of the process, type 'cfm-seriation f=/usr/share/cfm-seriation/data/Homo_sapiens.dat m=3000 P v' to execute the seriation. This will consume some extra time.
<pre>
Usage: cfm-seriation [OPTION...]
Seriation Parameters:
f=[NETWORK FILE].dat Network file path name
o=[ORDER FILE].dat Apply initial order
i=[INTERVAL] Number of isothermal steps
m=[STEPS] Number of steps
c=[FACTOR] Cooling factor
a=[ALPHA] Alpha value
p=[PERCENTUAL] Percentual energy for initial temperature
s=[SEDD] Random seed
P Plot graphs
v Generate video
Reading file...
Proteins: 9684
Interactions: 163509
Applying random order...
Saving and plotting initial order...
INITIAL Energy: 4123514310
Ordering...
100% [====================================================================================================]
</pre>
## Further Information
* [Optimization and Analysis of Seriation Algorithm for Ordering Protein Networks](ieeexplore.ieee.org/document/7033586/). This paper describes the optimizations implemented in this package.
* Felipe's Master Thesis [Otimização e análise de algoritmos de ordenamento de redes proteicas](http://hdl.handle.net/10923/6663). Full description of the optimizations implemented in this package (in Portuguese).
## License
The source code is distributed under the terms of the GNU General Public License v3 [GPL](http://www.gnu.org/copyleft/gpl.html).
## How to Cite this Package
If you are using this package on your research, please cite our paper:
* [Optimization and Analysis of Seriation Algorithm for Ordering Protein Networks](ieeexplore.ieee.org/document/7033586/)
<pre>
KUENTZER, Felipe A. et al. Optimization and analysis of seriation algorithm for ordering protein networks.
In: IEEE International Conference on Bioinformatics and Bioengineering (BIBE), 2014. p. 231-237.
</pre>
## Where Seriation is Used
If you are using the Seriation Package, please send an email to *alexandre.amory at pucrs.br* so we can update this list of users:
* [Transcriptogrammer](http://lief.if.ufrgs.br/pub/biosoftwares/transcriptogramer/)
## Similar Packages
* [R Package seriation](http://www.jstatsoft.org/v25/i03), available at http://cran.r-project.org/web/packages/seriation/index.html.
|